Research Projects: Genetics of Human Diseases
Research projects at the Tesson Laboratory are focused on the contribution of the genome in the development of heart disease, as well as on the analysis of the interactions between the genome and the environment as risk factors for cardiovascular conditions.Genetics of Heart Failure
Mutations in the lamin A/C gene (LMNA) cause a group of diseases called laminopathies of which 60% affect the heart (Dilated Cardiomyopathy [DCM], Atrial Fibrillation[AF]) and/or skeletal muscles (Emery-Dreifuss Muscular Dystrophy [EDMD]). The LMNA mutation-caused DCM results in very poor patient prognosis and is often present in individuals with EDMD. The AF, a form of arrhythmia which results in significant morbidity and mortality is also frequently seen in DCM and/or EDMD patients with heart conduction defects.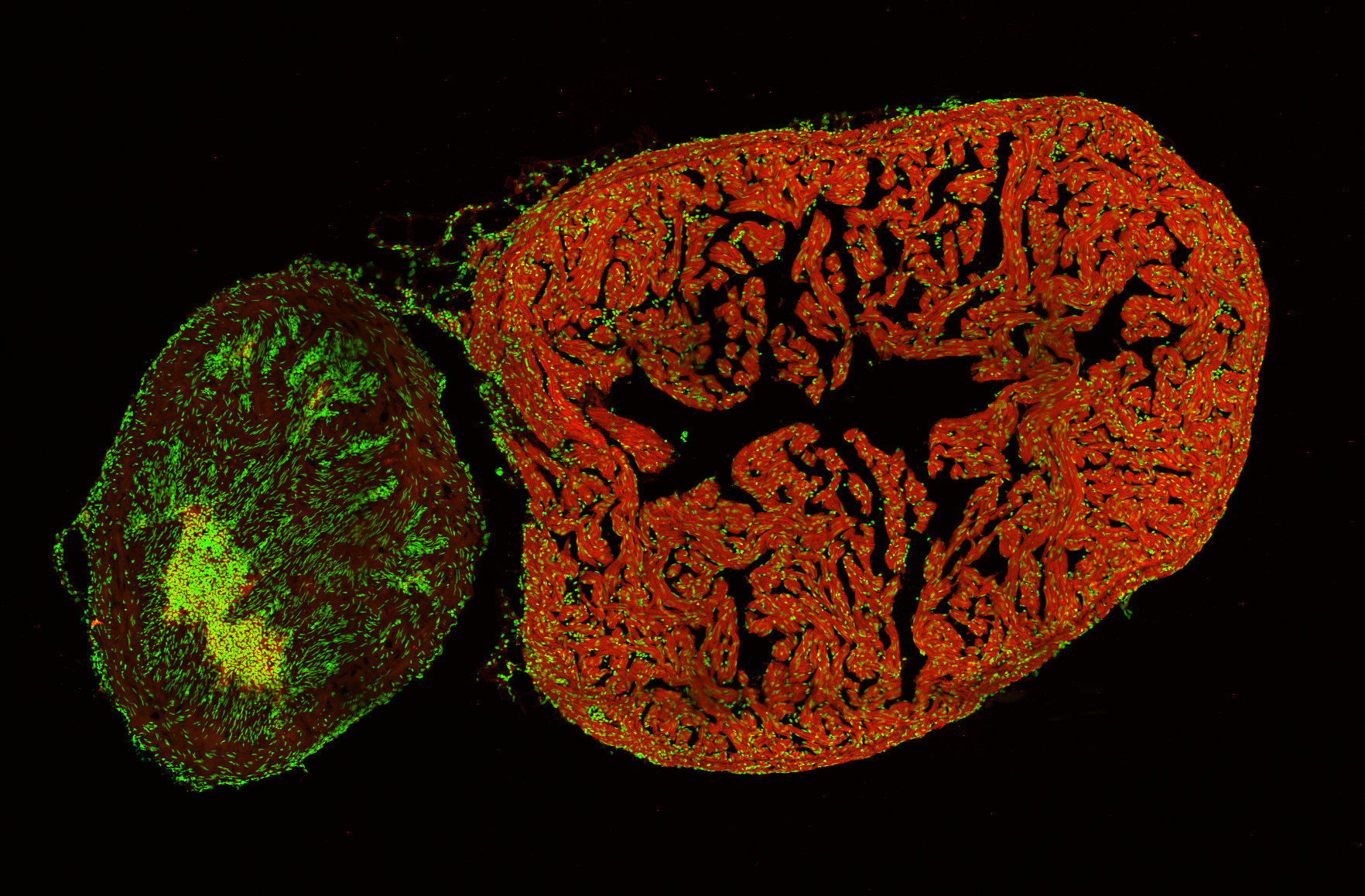
Zebrafish ventricle and bulbus arteriosus (cell nuclei in green, myosin heavy chain in red) by Dr. Hannah Nicolas
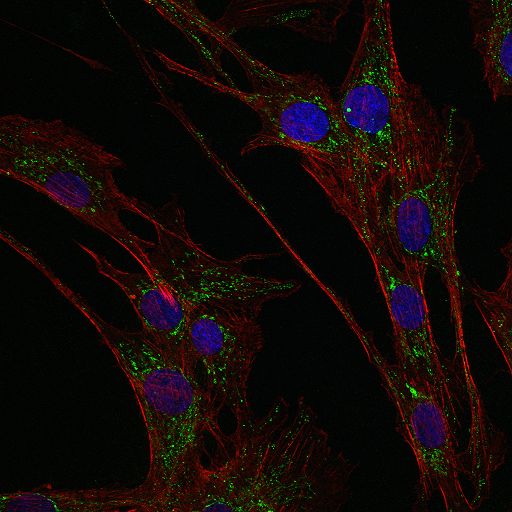
Human fibroblasts (cell nuclei in blue, F actin in red, HSP60 in green) by Dr. Hannah Nicolas
Overall Objectives:
- We first aim to study how mutations in an intranuclear structural protein, the lamin A/C, exert their cellular effect. To achieve this goal, we use LMNA transfected cell lines, mouse and human cell lines and tissues. This work is done in collaboration with Dr. Bilinska's clinical team at the Institute of Cardiology in Warsaw, Poland and Dr. Bonne's team at the Centre de Recherche en Myologie in Paris, France.
- We then aim to construct zebrafish models for the cardiac and skeletal muscle laminopathies. This will allow a better understanding of the physiopathological mechanisms underlying the diseases as well as to perform large-scale chemical compounds’ screens to evaluate their potential efficiency in rescuing the phenotype. This work is done in collaboration with Dr. Akimenko at the University of Ottawa.
Genetic Determinants of Weight Loss
Obesity and overweight are risk factors to many diseases including heart disease and cancer. The current projects focus on the environmental and genetic changes that influence the rate of weight loss in overweight individuals undergoing caloric restriction diets.Overall Objectives:
- Our previous studies showed the ACSL5 rs2419621 genotype to be associated with weight loss in a population of overweight women during a caloric restriction diet.
- The first objective is to determine the cellular and molecular mechanisms by which the genotype of the ACSL5 gene and other genes involved in fatty acid utilization in muscle cells exert their effect. This work is done in collaboration with Dr. Harper at the University of Ottawa and Dr. Prud’homme at the Université de Moncton.
- The second objective is to study the impact of the environmental toxicant MEHP (Mono(2-ethylhexyl) phthalate) on ACSL5 expression, fatty acid metabolism, and respiration in skeletal muscle. Found in food packaging, MEHP has been associated with obesity.
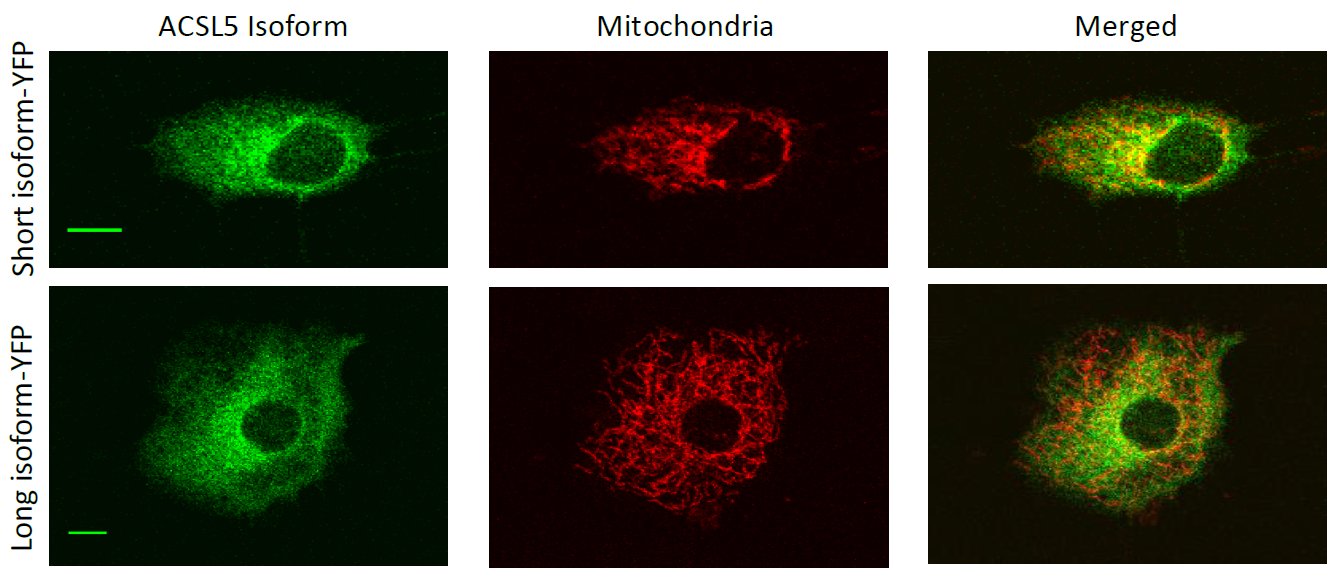
ACSL5 long and short human isoform-YFP (green) tagged colocalization in mitochondria (red) in C2C12 mouse myoblasts.
Genetic Determinants of Hypertension
High blood pressure is a condition that affects a large part of our population and is a key risk factor for cardiovascular mortality. The incidence of high blood pressure increases with age. About 50% of people with high blood pressure are salt sensitive, which means that blood pressure goes up with high salt consumption and drops when salt in the diet is restricted. There is a large body of evidence to support the strong contribution of our genetic make-up to salt sensitivity.Our database includes phenotypic characteristics and genotypic data for markers all along the genome of 605 individuals. Individuals were recruited at the Ottawa Heart Institute at the Hypertension clinic lead by Dr. Leenen as well as at the Institute of Cardiology in Warsaw (Poland) by the Hypertension clinic lead by Dr. Januszewicz. Samples were genotyped at 526,549 positions in the genome at the University of Ottawa Heart Institute.
Overall Objectives:
- We use whole-genome and whole-exome approaches to identifying genetic variants associated with salt-sensitive hypertension. Candidate genes have been selected based on genetic association and pathophysiological considerations. We have identified and continue to identify specific genetic make-up in these genes that are more frequent in salt sensitive individuals.
- We also aim to search the entire genome for chromosomal regions that are associated with salt-sensitivity. This unbiased approach might result in determination of new molecular pathways involved in blood pressure regulation.